Deskriptiv
Kvantitative data
Kategoriske data
Intervaller
Analytisk
Sandsynligheder
Kategoriske udfald
Kategoriske eksponeringerLogistisk regression
Kvantitative udfald
Kvantitative udfaldLinær regression
Korrelationer
Overlevelse
Poisson regression
Tilfældighed
Randomisering
Forskning
PhD thesis

Jacob Liljehult
Klinisk sygeplejespecialist
cand.scient.san, Ph.d.
Neurologisk afdeling
Nordsjællands Hospital
Linær regression
Simpel linær model
model1 <- lm(weight ~ age, data = strokedata)
summary(model1)
lm(formula = weight ~ age, data = apodata)
Residuals:
| Min | 1Q | Median | 3Q | Max |
|---|---|---|---|---|
| -40.138 | -12.137 | -1.291 | 10.901 | 102.555 |
| Estimate | Std. Error | t value | Pr(>|t|) | ||
|---|---|---|---|---|---|
| (Intercept) | 102.37581 | 3.17603 | 32.234 | <2e-16 | *** |
| age | -0.38471 | 0.04363 | -8.818 | <2e-16 | *** |
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Residual standard error: 16.87 on 981 degrees of freedom
(48 observations deleted due to missingness)
Multiple R-squared: 0.07344, Adjusted R-squared: 0.07249
F-statistic: 77.75 on 1 and 981 DF, p-value: < 2.2e-16
Justeret model
model2 <- lm(weight ~ age + height, data = strokedata)
summary(model2)
lm(formula = weight ~ age + height, data = strokedata)
Residuals:
| Min | 1Q | Median | 3Q | Max |
|---|---|---|---|---|
| -46.578 | -8.662 | -1.524 | 7.137 | 98.136 |
| Estimate | Std. Error | t value | Pr(>|t|) | ||
|---|---|---|---|---|---|
| (Intercept) | -79.61793 | 9.76465 | -8.154 | 1.08e-15 | *** |
| age | -0.15257 | 0.03902 | -3.910 | 9.88e-05 | *** |
| height | 0.96616 | 0.04982 | 19.394 | < 2e-16 | *** |
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Residual standard error: 14.36 on 978 degrees of freedom
(50 observations deleted due to missingness)
Multiple R-squared: 0.3307, Adjusted R-squared: 0.3293
F-statistic: 241.6 on 2 and 978 DF, p-value: < 2.2e-16
Goodness of fit
mf1 <- strokedata %>% select(age, weight, height) %>%
filter(!is.na(age) & !is.na(weight) & !is.na(height))
md1 <- lm(weight ~ age, data = mf1)
md2 <- lm(weight ~ age + height, data = mf1)
anova(md1,md2)
Model 1: weight ~ age
Model 2: weight ~ age + height
| Res.Df | RSS | Df | Sum of Sq | F | Pr(>F) | |||
|---|---|---|---|---|---|---|---|---|
| 1 | 979 | 279134 | ||||||
| 2 | 978 | 201599 | 1 | 77535 | 376.14 | <2.2e-16 | *** |
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Kontrol af modelantagelser
Kontrol af linearitet
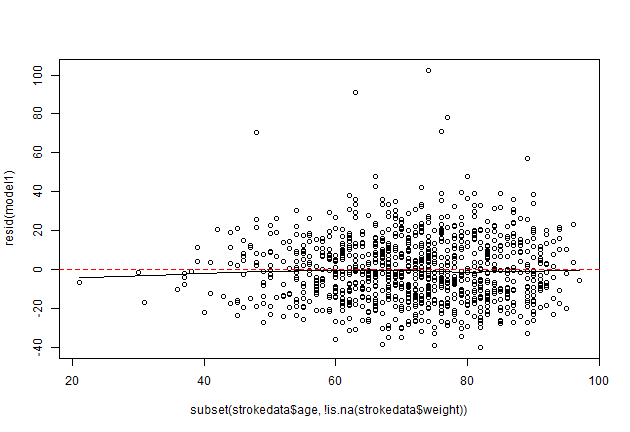
scatter.smooth(strokedata$age, resid(model1))
abline(0,0,col = "red", lty=2)
Error in xy.coords(x, y, xlabel, ylabel) : 'x' and 'y' lengths differ
scatter.smooth(subset(strokedata$age, !is.na(strokedata$weight)), resid(model1))
abline(0,0,col = "red", lty=2)
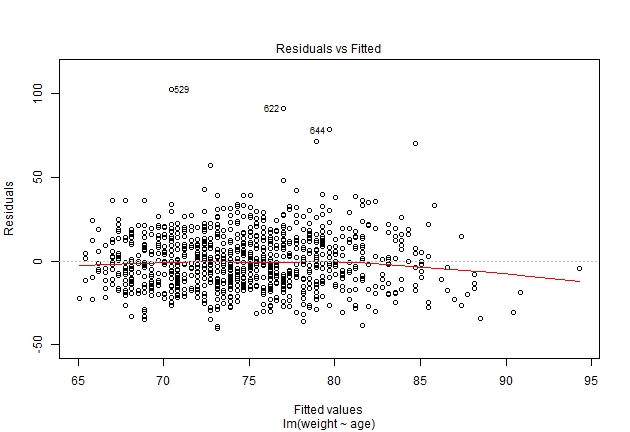
plot(model1, which = 1)
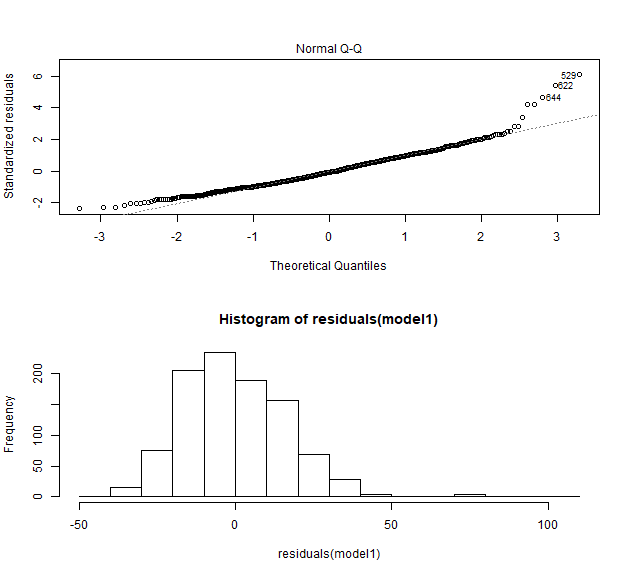
Normalfordeling af residualer
par(mfrow = c(2,1))
plot(model1, which = 2)
hist(residuals(model1))
Varianshomogenitet
plot(model1, which = 3)
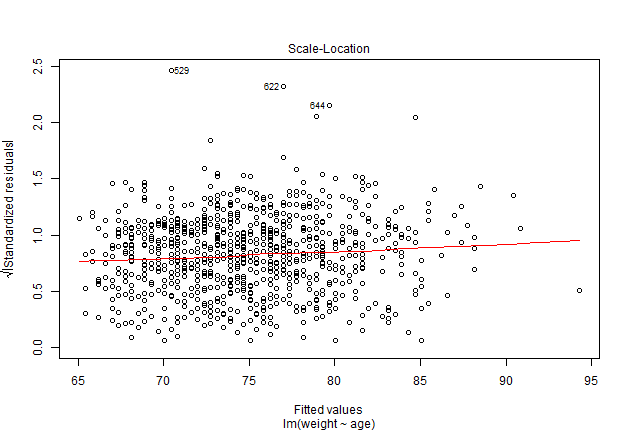
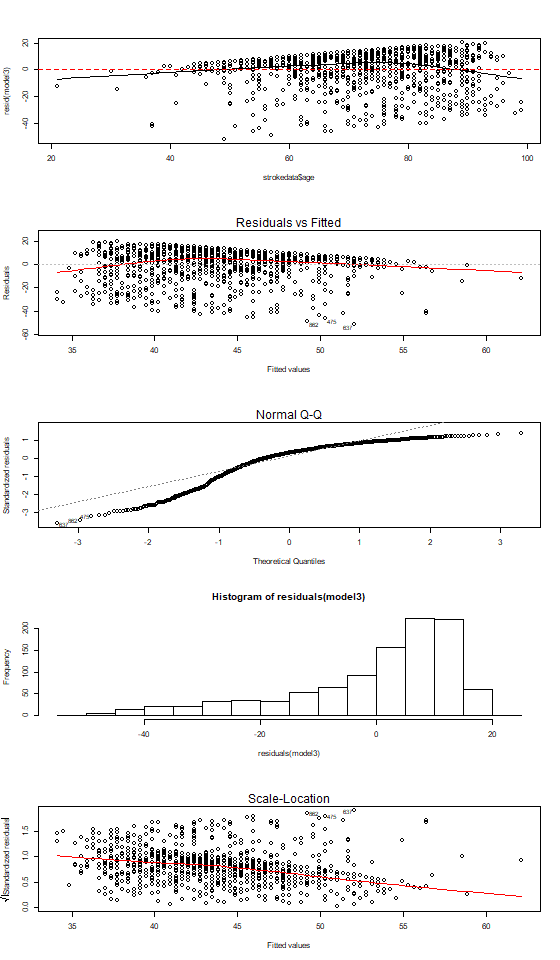
Eksempel på model hvor antagelserne ikke er opfyldt
model3 <- lm(sss ~ age, data = strokedata)
summary(model3)
lm(formula = sss ~ age, data = strokedata)
Residuals:
| Min | 1Q | Median | 3Q | Max | |
|---|---|---|---|---|---|
| -52.069 | -6.159 | 4.700 | 10.405 | 20.290 |
| Estimate | Std. Error | t value | Pr(>|t|) | ||
|---|---|---|---|---|---|
| (Intercept) | 69.66015 | 2.65238 | 26.263 | <2e-16 | *** |
| age | -0.35899 | 0.03638 | -9.869 | <2e-16 | *** |
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Residual standard error: 14.5 on 1029 degrees of freedom
Multiple R-squared: 0.08647, Adjusted R-squared: 0.08558
F-statistic: 97.4 on 1 and 1029 DF, p-value: < 2.2e-16
library(ggplot2)
ggplot(aes(x = age, y = sss), data = apodata) + geom_point() +
geom_smooth(method = loess) + theme_bw()
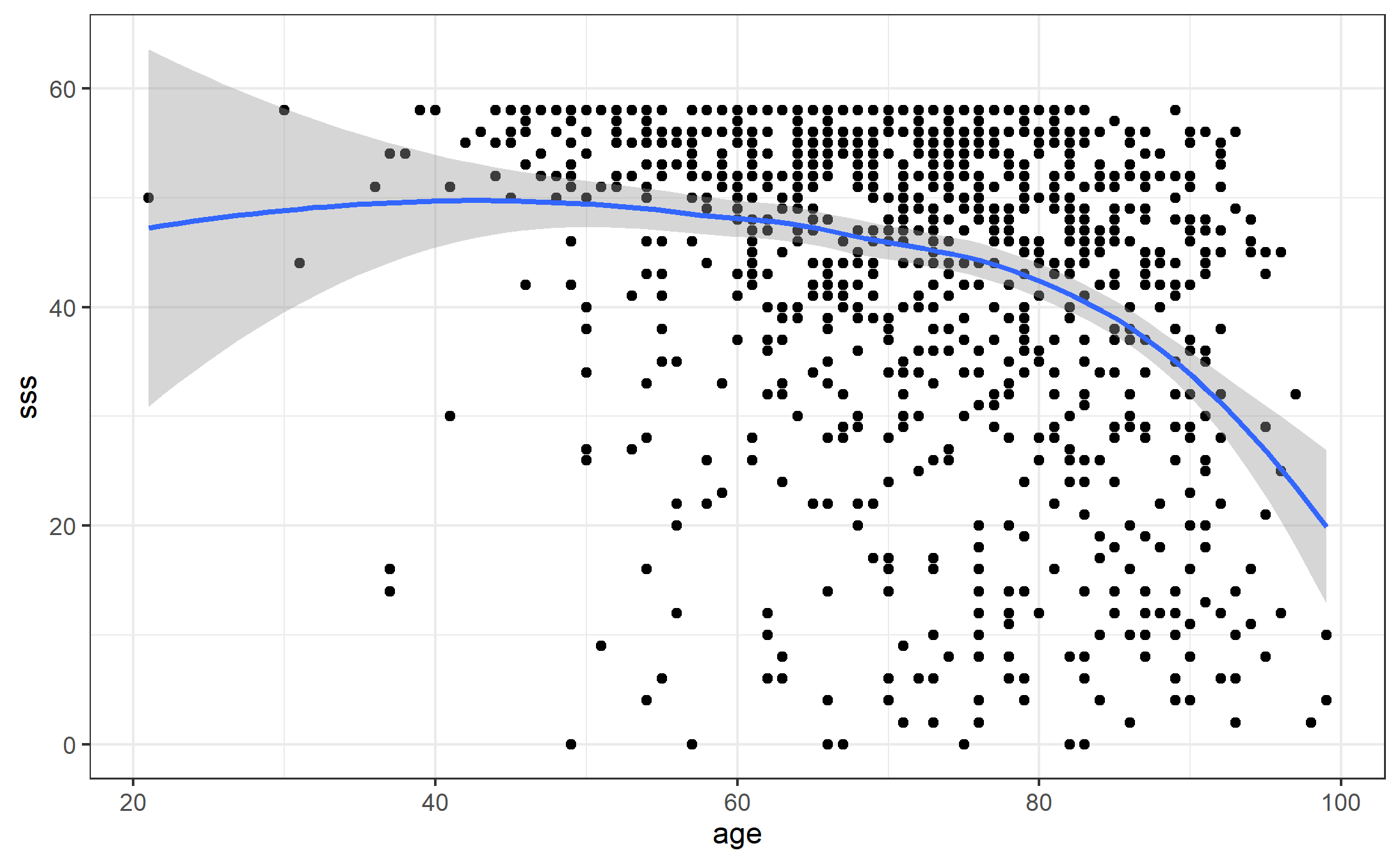
par(mfrow = c(5,1))
scatter.smooth(strokedata$age, resid(model3))
abline(0,0,col = "red", lty=2)
plot(model3, which = 1)
plot(model3, which = 2)
hist(residuals(model3))
plot(model3, which = 3)